Topic 4: LiverID - Molecular control of pathophysiological alterations to liver cell identities

Presentation of the research topic
Liver diseases are a major burden on human health. In particular, modern lifestyles, including excessive food and alcohol consumptions, are responsible for an ever-increasing number of liver failures and cancers.
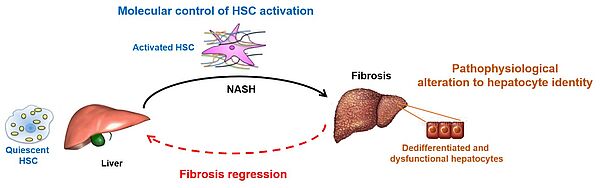
Liver diseases are caused by alterations in the activities of hepatocytes, the main cell type that makes up this organ. Indeed, a dysfunctional liver is characterized by unhealthy hepatocytes, unable to maintain normal homeostatic functions. Alterations to hepatocyte functions are accompanied by changes in the activities of other liver cell types, including hepatic stellate cells (HSCs). Indeed, HSCs are able to adopt a myofibroblastic phenotype that is responsible for the development of fibrosis, an advanced stage of chronic liver diseases characterized by an alteration of the extracellular matrix surrounding hepatocytes.
Our team studies the molecular mechanisms at play in diseased livers that underlie alterations in hepatocyte and HSC activities. Specifically, we investigate how alterations in gene expression occur in these cells and whether these can be corrected to restore normal cellular functions. Finally, given that liver cells communicate directly with each other, we are also studying how changes in gene expression translate into changes in the intercellular dialogue in diseased livers.
Through our work, we hope to contribute to a better understanding of the molecular mechanisms underlying alterations to liver functions with the aim to identify potential new strategies to treat liver diseases.
Our team studies the molecular mechanisms involved in the modulation of the transcriptome and phenotype of HSCs and hepatocytes, particularly in the context of NASH-associated fibrosis and its reversion.
Recent work from our team has allowed to:
- Identify sex and time as main variables impacting on transcriptional dysregulation in Human metabolic dysfunction-associated steatotic liver disease (MASLD) (Vandel et al. Hepatology. 2020; Johanns et al. JHEP Rep 2024)
- Unveil that hepatocyte identity is controlled by an extended array of transcription factors beyond the well-recognized core regulatory network including the thyroid hormone receptor beta (THRB) (Dubois-Chevalier et al. EMBO Rep 2023).
- Identify the transcription factor BNC2 and its o-GlcNacylation as cornerstones in establishment of the myofibroblastic transcriptional program involved in development of liver fibrosis (Bobowski-Gerard M et al. Nat Commun, 2022; Very N et al. Cell Death & Dis 2024)
- Demonstrates that seemingly reversible and unnoticed metabolic challenges in early adulthood may predispose the liver to exacerbated metabolic dysfunction when faced with chronic challenges later in life (Berthier et al. J Hepatol 2025)
- Characterize the epigenetic and transcriptional regulatory mechanisms underlying hepatocyte-to-cholangiocyte transdifferentiation (Vasseur et al., PLoS Biology 2025
Key-words
Bioinformatics ; cell identity ; circadian rythms ; fibrosis ; functional genomics ; gene transcriptional regulation ; liver pathophysiology ; nuclear receptors ; transcription factors
Jérôme EECKHOUTE
Research director 2, CNRS, team leader
ORCID : 0000-0002-7222-9264
Jérôme Eeckhoute earned his PhD in molecular biology from the University of Lille in 2003 owing to studies defining the functional consequences of diabetes-associated mutations of the nuclear receptor HNF4A. He subsequently moved to the Dana-Farber cancer institute (Harvard Medical School, Boston, USA) where he was involved in defining the transcription factor hierarchy and interplay with chromatin involved in regulatory activities of the estrogen receptor. Ever since his recruitment by the CNRS in 2008, he has been focusing on applying functional genomics to decipher molecular mechanisms involved in (dys)regulation of cell/tissue-specific transcriptional regulatory programs with a special emphasis on nuclear receptors and chromatin/epigenomics. Recent interests lie in the characterization of molecular entities and mechanisms responsible for establishment and maintenance/loss of cell identity in liver pathophysiology
Philippe LEFEBVRE
Research director 1, Inserm
ORCID : 0000-0002-9366-5129
Philippe Lefebvre completed his PhD in Biochemistry at ULille1 in 1988, during which he studied the mechanism of action of anti-glucocorticoids under the supervision of Pr P. Formstecher. He then did his postdoctoral training in Dr G.L. Hager’s lab at N.I.H., N.C.I., Bethesda to study transcriptional mechanisms governing the glucocorticoid-driven, cell type-specific expression of the Mouse Mammary Tumor Virus (MMTV) and chromatin-regulated processes. Since 1992, he has been at INSERM in Lille where he is currently a research director. He first established a research group working mostly on nuclear retinoic acid receptors (RARs, RXRs) and orphan receptors structure and regulation (Nurr1, Nur77) in cellular differentiation and cancer. His scientific interests then switched towards the study of the role of nuclear receptors in cardiometabolic diseases, mostly focusing on PPARα and FXR. He now leads a research group in Pr Bart Staels laboratory and, together with Dr Jérôme Eeckhoute, focuses mostly on nuclear receptor-regulated molecular mechanisms governing liver pathophysiology, with a specific emphasis on NASH and fibrosis.
Lora Autier
Engineer, Lille University
Lora Autier obtained a Master's degree in Biology-Health, specializing in Oncology, from the University of Lille in 2023. She then joined the CER Group, a biopharmaceutical company specializing in preclinical development and located in Belgium, where she participated in the development of analytical methods to evaluate immune responses, safety, and efficacy of biological products. In September 2025, she joined the research group headed by Dr. Jérôme Eeckhoute to study the transcriptional regulatory mechanisms controlling the identity and plasticity of liver cells in pathophysiological contexts.
Alexandre BERTHIER
Postdoctoral fellow
ORCID : 0000-0003-4153-4810
Alexandre Berthier earned his PhD in Life Sciences (molecular aspects) from the University of Besançon in 2008, where he developed "biosensors/cellular models" tools for identifying ligands of the nuclear estrogen receptor (ERα) and studying DNA/protein and protein/protein interactions. He subsequently investigated AMP-regulated kinase (AMPK) in Grenoble as part of Professor Uwe Schlattner’s group before joining Dr. Philippe Lefebvre’s team at the University of Lille in 2010. His research focuses on the nuclear receptors FXR, PPARα, and REV-ERBα, and their roles in the circadian regulation of liver metabolism. He is particularly interested in the transcriptional mechanisms involved in metabolic-associated steatohepatitis (MASH) and liver fibrosis, exploring the molecular pathways underlying these conditions. He develops innovative approaches to unravel complex molecular interactions and metabolic mechanisms.
Desiree de BRUIN
Ph.D Student, Lille University
ORCID : 0009-0005-4186-4627
Desiree de Bruin will joint the team in February 2025 as PhD candidate, part of the MSCA MIRACLE Doctoral Network. Her project focuses on liver cell plasticity in MASH related inflammation and fibrosis. Her background is in molecular biology (bachelor) and forensic science (master). She worked as a research technician at the Hubrecht Institute (Utrecht, The Netherlands) in the group of prof. Dr. Geert Kops, focusing on endogenous tagging of cell cycle regulating proteins using CRISPR techniques and live cell imaging. After completing her master’s, she worked as a research associate at the Amsterdam University Medical Center (Amsterdam, The Netherlands) under the supervision of Dr. Peter Henneman, focusing on forensic (epi)genetics and third generation sequencing techniques.
Julie DUBOIS-CHEVALIER
Research Engineer, INSERM, Bioinformatics platform leader
ORCID : 0000-0003-0471-752X
Julie Dubois-Chevalier is a specialist in Bioinformatics and Machine Learning. She earned her PhD in Chemoinformatics at the University of Orléans in 2011, studying the concepts of molecular “diversity” and “representativity” to propose a molecules selection method for high throughput screening in drug discovery. She joined the team in 2012 for a post-doctoral fellow under the supervision of Jérôme Eeckhoute to study the role of transcription factors and their cooperation in defining the genesis of the adipocyte lineage. Since 2016, she is bioinformatics platform leader for the unit, developing and deploying tools to process omics data. She leverages her expertise in Machine Learning to study the epigenetic mechanisms involved in the control of hepatocyte identity by integrating heterogeneous omics data.
Sandra COURQUET
Engineer, Lille University
ORCID : 0000-0002-2442-3222
Sandra Courquet obtained her master's degree in Biology and Health Products, Host-Graft Relation pathway at the University of Besançon in 2017. She then joined the CNR Echinococcoses/UMR6249 Chrono-Environnement (Besançon), where her work focused mainly on monitoring the Echinococcus multilocularis parasite, responsible for the liver disease Alveolar Echinococcosis. Then, in 2022, she joined the Clinical Research Department of Medical Oncology (Besançon University Hospital) to ensure the follow-up and quality of phase I to IV clinical trials. In October 2024, she joined UMR1011 under the direction of Dr Philippe Lefebvre, to participate in the study of molecular mechanisms involved in liver pathologies.
Hélène DEHONDT
Engineer, Lille University
ORCID : 0000-0003-2900-4673
Hélène Dehondt obtained her Master's degree in Cellular and Molecular Biology in 2000 at the University of Lille. She was then employed by the CNRS in Strasbourg to develop a new technique for discriminating mutations in colorectal cancer, and to manage a European research sub-project to highlight the toxicity of products derived from fatty acid ionization. Building on this experience, she joined UMR1011 in 2001. Her missions consist in two main areas: management of the technical platform and management of research projects aimed at understanding the molecular mechanisms involved in metabolic diseases.
Dmitry GALINOUSKY
Postdoctoral Fellow
ORCID : 0000-0002-2522-6052
Dmitry Galinousky is experienced in bioinformatics and data processing and is an expert in molecular genetics. Dmitry has developed approaches for predicting phenotypic traits based on omics data (transcriptomic, proteomic, and morphometric data). He has also examined the gut microbiome status of long-lived individuals. Dmitry joined the team in April 2024 thanks to a joined research program with the biopharmaceutical company Genfit. He is in charge of bulk and single-cell/nuclei RNA-seq data processing both using public and in-house datasets.
Céline GHEERAERT
Engineer, Lille University
ORCID : 0000-0003-4096-6363
Céline Gheeraert obtained a Master's degree in Cellular and Molecular Engineering from the University of Lille in 2006. She began her professional career at GENFIT, a biotechnology company based in Lille, where she contributed to projects focused on identifying novel pharmacological treatments for diseases associated with metabolic syndrome.
In 2008, she joined the unit under the supervision of Dr. Philippe Lefebvre as an engineer, where she developed expertise in cistrome analysis techniques and epigenomic modifications (ChIPseq, CUT&RUN, (h)MeDIPseq). She also set up and managed a gene expression microarray platform (Affymetrix technology) before evolving this technological approach towards the use of “new generation” sequencing (RNAseq/snRNAseq).
For several years, under the supervision of Dr. Jérôme Eeckhoute, she has focused on studying the transcriptional networks that regulate cell identity in liver pathophysiology. Her research particularly investigates the role of transcription factors in maintaining and modulating hepatic identity under both normal and pathological conditions.
Marie Lapage
Engineer, Lille University
Marie Lapage obtained a Master’s degree in Biotechnology, with a specialization in cellular and molecular engineering, from the University of Lille in 2024. She subsequently continued her training in the MIDAC laboratory, where she mainly worked on the evaluation of the virucidal and/or bactericidal efficacy of products. In January 2026, she joined UMR 1011 under the supervision of Dr. Philippe Lefebvre, to contribute to the study of molecular mechanisms involved in liver pathologies.
Georgiana TOMA
Postdoctoral fellow
Numéro ORCID : 0000-0003-0356-9158
Georgiana Toma received her PhD in Medical Immunology at the Martin-Luther University from Halle (Saale) in Germany. Her work was at the intersection of ageing, immunology and post-translational protein modifications, focusing on CD8+ T cells and lysine-acetylation patterns. She joined the team in 2022, shifting her focus to liver diseases and circadian rhythm. Her current work explores the interplay between glycolysis and the circadian clock genes in the context of liver fibrosis.
2026/01: New year celebration
2025/12: Ludivine Vasseur obtained her PhD ! Congratulations Ludivine !
2025/11: Two ANR PRC grants obtained by the LiverID team !
2025/10: Our team actively contributed to the “Fête de la Science”, engaging with elementary and middle school students to present the role of artificial intelligence and its connection to biology in our research.
2025/01: Céline Gheeraert promoted to the grade of “hors classe” engineer (University of Lille) ! Congratulations Céline on this well-deserved promotion!
Team 4 from UMR1011 developped lot of scientific skills :
- Mouse models of liver injury/diseases; PCLS; Primary mouse liver cells (hepatocytes, HSC…)
- Experimental analyses of gene transcriptional regulation through targeted and multi-omics appraoches (bulk and scRNAseq; ChIPseq; RIME etc)
- Bioinformatical data mining and Machine Learning for Data Integration
List of main publications from Team 4:
Derepression of the epithelial transcription factor GRHL2 promotes direct hepatocyte-to-cholangiocyte transdifferentiation. Vasseur L, Gheeraert C, Dubois-Chevalier J, Very N, Guille L, Bou Saleh M, Boulet C, Sobolewski C, Loyer P, Berthier A, Legrand N, Corlu A, Gnemmi V, Lasailly G, Leteurtre E, Galinousky D, Bongiovanni A, Taront S, Toft NI, Grøntved L, Tulasne D, Furlan A, Ntandja-Wandji LC, Staels B, Lefebvre P, Dubuquoy L, Eeckhoute J. PLoS Biol. 2025 Dec 12;23(12):e3003547. doi: 10.1371/journal.pbio.3003547
https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3003547
Unveiling the Molecular Legacy of Transient Insulin Resistance: Implications for Hepatic Metabolic Adaptability. Berthier A, Gheeraert C, Vinod M, Johanns M, Guille L, Haas JT, Dubois-Chevalier J, Eeckhoute J, Staels B, Lefebvre P. J Hepatol. 2025 Feb 11:S0168-8278(25)00080-7.
https://www.journal-of-hepatology.eu/article/S0168-8278(25)00080-7/fulltext
Increased O-GlcNAcylation connects metabolic to transcriptional reprogramming during pathophysiological cell activation. Very N, Staels B, Eeckhoute J. Trends Cell Biol. 2024 Nov 7:S0962-8924(24)00213-7.
https://pubmed.ncbi.nlm.nih.gov/39516052/
O-GlcNAcylation controls pro-fibrotic transcriptional regulatory signaling in myofibroblasts. Very N, Boulet C, Gheeraert C, Berthier A, Johanns M, Bou Saleh M, Guille L, Bray F, Strub JM, Bobowski-Gerard M, Zummo FP, Vallez E, Molendi-Coste O, Woitrain E, Cianférani S, Montaigne D, Ntandja-Wandji LC, Dubuquoy L, Dubois-Chevalier J, Staels B, Lefebvre P, Eeckhoute J.Cell Death Dis. 2024 Jun 3;15(6):391.
https://www.nature.com/articles/s41419-024-06773-9
Nuclear receptors: pathophysiological mechanisms and drug targets in liver disease. Dubois V, Lefebvre P, Staels B, Eeckhoute J. Gut 2024 Jun 11:gutjnl-2023-331741.
https://gut.bmj.com/content/early/2024/06/11/gutjnl-2023-331741.long
The Molecular Circadian Clock Is a Target of Anti-cancer Translation Inhibitors. Berthier A, Gheeraert C, Johanns M, Vinod M, Staels B, Eeckhoute J, Lefebvre P. J Biol Rhythms. 2024 Feb;39(1):20-34.
https://hal.science/hal-04264765
Time-of-day-dependent variation of the human liver transcriptome and metabolome is disrupted in MASLD. Johanns M, Haas JT, Raverdy V, Vandel J, Chevalier-Dubois J, Guille L, Derudas B, Legendre B, Caiazzo R, Verkindt H, Gnemmi V, Leteurtre E, Derhourhi M, Bonnefond A, Froguel P, Eeckhoute J, Lassailly G, Mathurin P, Pattou F, Staels B, Lefebvre P. JHEP Rep. 2023 Oct 27;6(1):100948.
https://www.jhep-reports.eu/article/S2589-5559(23)00279-3/fulltext
An extended transcription factor regulatory network controls hepatocyte identity. Dubois-Chevalier J, Gheeraert C, Berthier A, Boulet C, Dubois V, Guille L, Fourcot M, Marot G, Gauthier K, Dubuquoy L, Staels B, Lefebvre P, Eeckhoute J. EMBO rep. 2023 Jul 10:e57020.
https://www.embopress.org/doi/full/10.15252/embr.202357020
A time- and space-resolved nuclear receptor atlas in mouse liver. Zummo FP, Berthier A, Gheeraert C, Vinod M, Bobowski-Gérard M, Molendi-Coste O, Pineau L, Jung M, Guille L, Chevalier-Dubois J, Dombrowicz D, Staels B, Eeckhoute J, Lefebvre P. J Mol Endocrinol. 2023 Mar 1:JME-23-0017.
https://pubmed.ncbi.nlm.nih.gov/36988391/
Functional genomics uncovers the transcription factor BNC2 as required for myofibroblastic activation in fibrosis. Bobowski-Gerard M, Boulet C, Zummo PF, Dubois-Chevalier J, Gheeraert C, Bou Saleh M, Strub J-M, Farce A, Ploton M, Guille L, Vandel J, Bongiovanni A, Very N, Woitrain E, Deprince A, Lalloyer F, Bauge E, Ferri L, Ntandja-Wandji L-C, Cotte KA, Grangette C, Vallez E, Cianférani S, Raverdy V, Caiazzo R, Gnemmi V, Leteurtre E, Pourcet B, Paumelle R, Ravnskjaer K, Lassailly G, Haas TJ, Mathurin P, Pattou F, Dubuquoy L, Staels B, Lefebvre P, Eeckhoute J.. Nat Commun, 2022 Sep 10;13(1):5324.
https://www.nature.com/articles/s41467-022-33063-9
Timed use of digoxin prevents heart ischemia–reperfusion injury through a REV-ERBα–UPS signaling pathway. Vinod M, Berthier A, Maréchal X, Gheeraert C, Boutry R, Delhaye S, Annicotte J-S, Duez H, Hovasse A, Cianférani S, Montaigne D, Eeckhoute J, Staels B, Lefebvre P.Nature Cardiovascular Research, 2022
https://www.nature.com/articles/s44161-022-00148-z
Hepatic molecular signatures highlight the Non-Alcoholic SteatoHepatitis (NASH) sexual dimorphism. Vandel J, Dubois-Chevalier J, Gheeraert C, Derudas B, Raverdy V, Thuillier D, Van Gaal L, Francque S, Pattou F, Staels B, Eeckhoute J, Lefebvre P. Hepatology. 2020 Mar;73(3):920-936. doi: 10.1002/hep.31312.
https://pubmed.ncbi.nlm.nih.gov/32394476/
The Nuclear Bile Acid Receptor FXR is a PKA- and FOXA2-Sensitive Activator of Fasting Hepatic Gluconeogenesis. Ploton M, Mazuy C, Gheeraert C, Dubois V, Berthier A, Dubois-Chevalier J, Maréchal X, Bantubungi-Blum K, Diemer H, Cianférani S, Strub JM, Helleboid-Chapman A, Eeckhoute J, Staels B, Lefebvre P. J Hepatol. 2018 Jul 4. pii: S0168-8278(18)32175-5.
https://linkinghub.elsevier.com/retrieve/pii/S0168-8278(18)32175-5
Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV-ERBα complex. Berthier A, Vinod M, Porez G, Steenackers A, Alexandre J, Yamakawa N, Gheeraert C, Ploton M, Maréchal X, Dubois-Chevalier J, Hovasse A, Schaffer-Reiss C, Cianférani S, Rolando C, Bray F, Duez H, Eeckhoute J, Lefebvre T, Staels B, Lefebvre P. Proc Natl Acad Sci U S A. 2018 Nov 5. pii: 201805397.
https://pubmed.ncbi.nlm.nih.gov/30397120/
List of fundings obtained by Team 4 :

A cornerstone of our team’s strategy is the integration of wet- and dry-lab work. Our projects engage both contractual and permanent staff members, leveraging their complementary expertise in these areas.
We welcome Master students, PhD candidates, and Post-docs with an interest in functional genomics and liver pathophysiology. Our team includes both biologists and bioinformaticians, fostering a collaborative environment.
If you are interested in joining us, please contact:
Dr Jérôme Eeckhoute (jerome.eeckhoute[at]inserm.fr)
Dr Jérôme Eeckhoute
Research director, CNRS
jerome.eeckhoute[at]inserm.fr
Postal adress : Boulevard du Pr Jules Leclercq, Batiment J&K -Faculté de médecine - Pôle Recherche, 59045 Lille Cedex
Dr Philippe Lefebvre
Research director, INSERM
philippe-claude.lefebvre[at]inserm.fr
Postal adress : Boulevard du Pr Jules Leclercq, Batiment J&K -Faculté de médecine - Pôle Recherche, 59045 Lille Cedex
Dubois Vanessa (Post-doc) - Prof endocrinology and team leader, Ghent University
Boulet Clémence (IE)
Bobowski-Gerard Marie (Post-doc) - R&D project leader, Genfit
Vandel Jimmy (Post-doc) - CNRS research engineer
Zummo Francesco (Post-doc) - Medical advisor, Novo Nordisk
Guille Loic (IE) - Bioinformatics engineer, UC Louvain
Johanns Manuel (Post-doc) - Post-doc, UC Louvain
Vinod Manjula (Post-doc) - Senior scientist, Genfit
Avilkina Viktorija (Post-doc) - R&D Project Management, CisBio (Revvity)
Very Ninon (Post-doc) - Post-doc, University of Lille
Vasseur Ludivine (PhD) - ATER, University of Amiens



